Table manipulation#
Pandas, short for Python Data Analysis Library, is a modern, powerful and feature rich library that is designed for doing data analysis in Python. Here, we will cover some basic Pandas that will allow you to store results in table format, do some basic table operations, visualization, and to read and write tables to files. You will find more information online in the Pandas Tutorials and in chapter 3 of the Python Data Science Handbook [VanderPlas, 2016].
import numpy as np
import pandas as pd
pd.__version__
'2.3.3'
DataFrame#
The main pandas data structure is a table format called DataFrame. A DataFrame contains an Index and it contains columns. Columns are one-dimensional arrays of a single type called Series. A DataFrame can be created from a:
single Series object
list of dicts
dictionary of Series objects
two-dimensional NumPy array
NumPy structured array
mySeries = pd.Series(['London', 'Paris', 'Berlin'], name='City', dtype='string')
cities = pd.DataFrame({"City": mySeries, "Population": [8.8, 2.2, 3.6]})
cities
| City | Population | |
|---|---|---|
| 0 | London | 8.8 |
| 1 | Paris | 2.2 |
| 2 | Berlin | 3.6 |
DataFrames have the following attributes: dtypes, shape, index, columns, values, empty.
cities.columns
Index(['City', 'Population'], dtype='object')
cities.dtypes
City string[python]
Population float64
dtype: object
cities.shape
(3, 2)
cities.values
array([['London', 8.8],
['Paris', 2.2],
['Berlin', 3.6]], dtype=object)
Reading and writing#
The pandas library can read a variety of data formats using pandas.read_*(), including CSV, JSON, HTML, HDF5, Excel, SQL, SPSS, SAS, etc.:
fname = '../../data/airquality.csv'
airq = pd.read_csv(fname, index_col=0)
airq.head()
| Ozone | Solar | Wind | Temp | Month | Day | |
|---|---|---|---|---|---|---|
| ID | ||||||
| 101 | 41.0 | 190.0 | 7.4 | 67 | 5 | 1 |
| 102 | 36.0 | 118.0 | 8.0 | 72 | 5 | 2 |
| 103 | 12.0 | 149.0 | 12.6 | 74 | 5 | 3 |
| 104 | 18.0 | 313.0 | 11.5 | 62 | 5 | 4 |
| 105 | NaN | NaN | 14.3 | 56 | 5 | 5 |
The DataFrame object contains to_*() methods to write it to a file or database, e.g. airq.to_csv(out_filename).
Inspect DataFrame#
DataFrames have several methods that are useful for inspection: head(), tail(), and describe()
airq.describe()
| Ozone | Solar | Wind | Temp | Month | Day | |
|---|---|---|---|---|---|---|
| count | 116.000000 | 146.000000 | 153.000000 | 153.000000 | 153.000000 | 153.000000 |
| mean | 42.129310 | 185.931507 | 9.957516 | 77.882353 | 6.993464 | 15.803922 |
| std | 32.987885 | 90.058422 | 3.523001 | 9.465270 | 1.416522 | 8.864520 |
| min | 1.000000 | 7.000000 | 1.700000 | 56.000000 | 5.000000 | 1.000000 |
| 25% | 18.000000 | 115.750000 | 7.400000 | 72.000000 | 6.000000 | 8.000000 |
| 50% | 31.500000 | 205.000000 | 9.700000 | 79.000000 | 7.000000 | 16.000000 |
| 75% | 63.250000 | 258.750000 | 11.500000 | 85.000000 | 8.000000 | 23.000000 |
| max | 168.000000 | 334.000000 | 20.700000 | 97.000000 | 9.000000 | 31.000000 |
Select columns#
airq.Ozone
ID
101 41.0
102 36.0
103 12.0
104 18.0
105 NaN
...
249 30.0
250 NaN
251 14.0
252 18.0
253 20.0
Name: Ozone, Length: 153, dtype: float64
…or using a name or a list of names.
airq["Ozone"].head()
ID
101 41.0
102 36.0
103 12.0
104 18.0
105 NaN
Name: Ozone, dtype: float64
airq[["Ozone", "Solar"]].head()
| Ozone | Solar | |
|---|---|---|
| ID | ||
| 101 | 41.0 | 190.0 |
| 102 | 36.0 | 118.0 |
| 103 | 12.0 | 149.0 |
| 104 | 18.0 | 313.0 |
| 105 | NaN | NaN |
Asign new column#
airq["logOzone"] = np.log(airq.Ozone)
airq.head()
| Ozone | Solar | Wind | Temp | Month | Day | logOzone | |
|---|---|---|---|---|---|---|---|
| ID | |||||||
| 101 | 41.0 | 190.0 | 7.4 | 67 | 5 | 1 | 3.713572 |
| 102 | 36.0 | 118.0 | 8.0 | 72 | 5 | 2 | 3.583519 |
| 103 | 12.0 | 149.0 | 12.6 | 74 | 5 | 3 | 2.484907 |
| 104 | 18.0 | 313.0 | 11.5 | 62 | 5 | 4 | 2.890372 |
| 105 | NaN | NaN | 14.3 | 56 | 5 | 5 | NaN |
Select rows#
Slicing#
Slicing in pandas lets you select a continuous range of rows using start and end indices. It follows Python’s slicing rules, meaning it uses integer positions and the end index is exclusive. This provides a quick way to grab a block of consecutive rows. However, this syntax can be confusing because df[...] is also used for column selection, so airq[2:4] may look like it’s selecting columns when it’s actually slicing rows. Using .iloc is clearer, since it explicitly signals positional row access and avoids this ambiguity.
airq[2:4]
| Ozone | Solar | Wind | Temp | Month | Day | logOzone | |
|---|---|---|---|---|---|---|---|
| ID | |||||||
| 103 | 12.0 | 149.0 | 12.6 | 74 | 5 | 3 | 2.484907 |
| 104 | 18.0 | 313.0 | 11.5 | 62 | 5 | 4 | 2.890372 |
Boolean indexing#
Boolean indexing in pandas selects rows based on a condition that evaluates to True or False for each row. It allows filtering data according to one or more logical expressions and supports combining conditions with logical operators (&, |, and ~). This method is flexible and one of the most commonly used ways to subset DataFrames.
Note: Under the hood, boolean indexing works by creating a mask of True and False values that determines which rows are selected.
airq[(airq.Ozone > 15) & (airq.Ozone < 18)]
| Ozone | Solar | Wind | Temp | Month | Day | logOzone | |
|---|---|---|---|---|---|---|---|
| ID | |||||||
| 112 | 16.0 | 256.0 | 9.7 | 69 | 5 | 12 | 2.772589 |
| 182 | 16.0 | 7.0 | 6.9 | 74 | 7 | 21 | 2.772589 |
| 195 | 16.0 | 77.0 | 7.4 | 82 | 8 | 3 | 2.772589 |
| 243 | 16.0 | 201.0 | 8.0 | 82 | 9 | 20 | 2.772589 |
Query#
The query() method in pandas provides a way to filter rows using a string expression, making complex conditions more readable and concise. It can handle multiple conditions and logical operators, and even allows referencing variables from the environment using @.
month = 5
airq.query("Month == @month and Day < 5")
| Ozone | Solar | Wind | Temp | Month | Day | logOzone | |
|---|---|---|---|---|---|---|---|
| ID | |||||||
| 101 | 41.0 | 190.0 | 7.4 | 67 | 5 | 1 | 3.713572 |
| 102 | 36.0 | 118.0 | 8.0 | 72 | 5 | 2 | 3.583519 |
| 103 | 12.0 | 149.0 | 12.6 | 74 | 5 | 3 | 2.484907 |
| 104 | 18.0 | 313.0 | 11.5 | 62 | 5 | 4 | 2.890372 |
Select rows and columns#
Unlike the previous selection methods, .loc and .iloc can select both rows and columns at the same time. .loc is label-based, meaning you select rows and columns using their index labels or column names. .iloc is position-based, meaning you select rows and columns by integer positions.
Using the iloc indexer, we can index the underlying array as if it is a simple NumPy array (using the implicit Python-style index), but the DataFrame index and column labels are maintained in the result:
airq.iloc[1:3, 2:4]
| Wind | Temp | |
|---|---|---|
| ID | ||
| 102 | 8.0 | 72 |
| 103 | 12.6 | 74 |
Similarly, using the loc indexer we can index the underlying data in an array-like style but using the explicit row index and column indices:
airq.loc[100:102, ["Wind", "Temp"]]
| Wind | Temp | |
|---|---|---|
| ID | ||
| 101 | 7.4 | 67 |
| 102 | 8.0 | 72 |
Chained indexing#
To extract rows and columns, we may chain the commands pulling the columns and then the rows or vice versa (order does not matter). However, we should avoid chained indexing as it can lead to unpredictable results and warnings. Instead, we should use .loc or .iloc for clarity and reliability.
The expression below is an example of chained indexing. First, airq[["Ozone", "Solar"]] selects the two columns, creating a new temporary DataFrame (See View vs Copy ). Then, [airq.Ozone > 130] tries to filter rows on that temporary DataFrame using a condition from the original DataFrame.
The problem with this approach is that it can lead to unexpected behavior or warnings, because pandas is unsure whether the row filtering is applied to a view of the original DataFrame or a copy. This can cause issues if you later try to modify the filtered DataFrame.
airq["Ozone"][airq.Ozone > 130] = 130
/var/folders/d6/_vs0dtkd76d27wvhhz0_nn7h0000gn/T/ipykernel_44437/4132742754.py:1: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
airq["Ozone"][airq.Ozone > 130] = 130
A safer alternative is to use .loc, which avoids chaining:
airq.loc[airq.Ozone > 130, "Ozone"] = 130
Methods#
You can do masking and other useful things directly via methods of Series and DataFrames., e.g.
airq.Ozone.between(15, 18)
airq.Month.isin([5, 8])
airq.Month.astype(“string”)
airq.isna().sum()
Ozone 37
Solar 7
Wind 0
Temp 0
Month 0
Day 0
logOzone 37
dtype: int64
Simple aggregation functions include mean(), median(), min(), max(), and std().
airq['Ozone'].mean()
np.float64(41.758620689655174)
Grouping#
Sometimes we want to apply aggregation functions for individual factors separately.
airq.groupby('Month')
<pandas.core.groupby.generic.DataFrameGroupBy object at 0x119914980>
Sometimes we want to apply aggregation functions for individual factors separately.
airq.groupby('Month').mean()
| Ozone | Solar | Wind | Temp | Day | logOzone | |
|---|---|---|---|---|---|---|
| Month | ||||||
| 5 | 23.615385 | 181.296296 | 11.622581 | 65.548387 | 16.0 | 2.812076 |
| 6 | 29.444444 | 190.166667 | 10.266667 | 79.100000 | 15.5 | 3.236673 |
| 7 | 58.923077 | 216.483871 | 8.941935 | 83.903226 | 16.0 | 3.883834 |
| 8 | 58.500000 | 171.857143 | 8.793548 | 83.967742 | 16.0 | 3.845345 |
| 9 | 31.448276 | 167.433333 | 10.180000 | 76.900000 | 15.5 | 3.218795 |
Groupby comes with additional aggregation methods that provide more flexibility: aggregate(), filter(), transform(), and apply().
airq.groupby('Month').aggregate(["min", "max", "mean"])
| Ozone | Solar | Wind | Temp | Day | logOzone | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| min | max | mean | min | max | mean | min | max | mean | min | max | mean | min | max | mean | min | max | mean | |
| Month | ||||||||||||||||||
| 5 | 1.0 | 115.0 | 23.615385 | 8.0 | 334.0 | 181.296296 | 5.7 | 20.1 | 11.622581 | 56 | 81 | 65.548387 | 1 | 31 | 16.0 | 0.000000 | 4.744932 | 2.812076 |
| 6 | 12.0 | 71.0 | 29.444444 | 31.0 | 332.0 | 190.166667 | 1.7 | 20.7 | 10.266667 | 65 | 93 | 79.100000 | 1 | 30 | 15.5 | 2.484907 | 4.262680 | 3.236673 |
| 7 | 7.0 | 130.0 | 58.923077 | 7.0 | 314.0 | 216.483871 | 4.1 | 14.9 | 8.941935 | 73 | 92 | 83.903226 | 1 | 31 | 16.0 | 1.945910 | 4.905275 | 3.883834 |
| 8 | 9.0 | 130.0 | 58.500000 | 24.0 | 273.0 | 171.857143 | 2.3 | 15.5 | 8.793548 | 72 | 97 | 83.967742 | 1 | 31 | 16.0 | 2.197225 | 5.123964 | 3.845345 |
| 9 | 7.0 | 96.0 | 31.448276 | 14.0 | 259.0 | 167.433333 | 2.8 | 16.6 | 10.180000 | 63 | 93 | 76.900000 | 1 | 30 | 15.5 | 1.945910 | 4.564348 | 3.218795 |
Append#
Append rows#
The stack two data.frames together by row, use the concat() function. Note append() will be depreciated in future versions of pandas.
pd.concat([airq.head(2), airq.tail(2)])
| Ozone | Solar | Wind | Temp | Month | Day | logOzone | |
|---|---|---|---|---|---|---|---|
| ID | |||||||
| 101 | 41.0 | 190.0 | 7.4 | 67 | 5 | 1 | 3.713572 |
| 102 | 36.0 | 118.0 | 8.0 | 72 | 5 | 2 | 3.583519 |
| 252 | 18.0 | 131.0 | 8.0 | 76 | 9 | 29 | 2.890372 |
| 253 | 20.0 | 223.0 | 11.5 | 68 | 9 | 30 | 2.995732 |
Append columns#
The concat() function can also be used to stack columns of DataFrames together. The row index is preserved with an outer join by default.
pd.concat([airq.head(2), airq.tail(2)], axis=1)
| Ozone | Solar | Wind | Temp | Month | Day | logOzone | Ozone | Solar | Wind | Temp | Month | Day | logOzone | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ID | ||||||||||||||
| 101 | 41.0 | 190.0 | 7.4 | 67.0 | 5.0 | 1.0 | 3.713572 | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 102 | 36.0 | 118.0 | 8.0 | 72.0 | 5.0 | 2.0 | 3.583519 | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 252 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | 18.0 | 131.0 | 8.0 | 76.0 | 9.0 | 29.0 | 2.890372 |
| 253 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | 20.0 | 223.0 | 11.5 | 68.0 | 9.0 | 30.0 | 2.995732 |
To ignore row indices when stacking columns, you need to clear them prior to calling pd.concat().
df1 = airq.head(2).copy().reset_index(drop=True)
df2 = airq.tail(2).copy().reset_index(drop=True)
pd.concat([df1, df2], axis=1)
| Ozone | Solar | Wind | Temp | Month | Day | logOzone | Ozone | Solar | Wind | Temp | Month | Day | logOzone | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 41.0 | 190.0 | 7.4 | 67 | 5 | 1 | 3.713572 | 18.0 | 131.0 | 8.0 | 76 | 9 | 29 | 2.890372 |
| 1 | 36.0 | 118.0 | 8.0 | 72 | 5 | 2 | 3.583519 | 20.0 | 223.0 | 11.5 | 68 | 9 | 30 | 2.995732 |
Merge#
Pandas implements several ways of combining datasets via the pd.merge() function and the join()methods of Series and DataFrames.
The pd.merge() function implements one-to-one, many-to-one, and many-to-many joins.
The pd.merge() function joins based on column names and not the Index. The column names can be explicitly defined via the keywords on, left_on, and right_on.
View vs Copy#
When you create a new DataFrame by slicing an existing one, pandas often returns a view, not an independent copy. As a result, any modifications you make to that sliced DataFrame can unexpectedly alter the original data. To avoid accidental changes, explicitly create a copy when needed.
airq_view = airq.query("Month == 5 and Day < 5")
airq_view["Ozone"] = 0
airq.query("Month == 5 and Day < 5")
/var/folders/d6/_vs0dtkd76d27wvhhz0_nn7h0000gn/T/ipykernel_44437/3954834801.py:2: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame.
Try using .loc[row_indexer,col_indexer] = value instead
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
airq_view["Ozone"] = 0
| Ozone | Solar | Wind | Temp | Month | Day | logOzone | |
|---|---|---|---|---|---|---|---|
| ID | |||||||
| 101 | 41.0 | 190.0 | 7.4 | 67 | 5 | 1 | 3.713572 |
| 102 | 36.0 | 118.0 | 8.0 | 72 | 5 | 2 | 3.583519 |
| 103 | 12.0 | 149.0 | 12.6 | 74 | 5 | 3 | 2.484907 |
| 104 | 18.0 | 313.0 | 11.5 | 62 | 5 | 4 | 2.890372 |
To avoid accidental changes, explicitly create a copy when needed.
airq_copy = airq.query("Month == 5 and Day < 5").copy()
airq_copy["Ozone"] = 0
airq_copy.query("Month == 5 and Day < 5")
| Ozone | Solar | Wind | Temp | Month | Day | logOzone | |
|---|---|---|---|---|---|---|---|
| ID | |||||||
| 101 | 0 | 190.0 | 7.4 | 67 | 5 | 1 | 3.713572 |
| 102 | 0 | 118.0 | 8.0 | 72 | 5 | 2 | 3.583519 |
| 103 | 0 | 149.0 | 12.6 | 74 | 5 | 3 | 2.484907 |
| 104 | 0 | 313.0 | 11.5 | 62 | 5 | 4 | 2.890372 |
Visualization#
matplotlib#
%matplotlib inline
import matplotlib.pyplot as plt
plt.plot(airq.Ozone, airq.Temp, 'or')
plt.show()
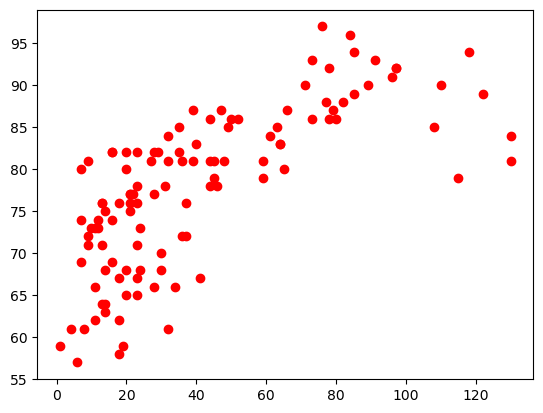
Pandas#
Pandas provides visualization tools built on top of matplotlib. Hence, you still need to import matplotlib’s pyplot. You can read more in the pandas reference guide here. There are two ways to use pandas for plotting:
via
plot()methods for Series and DataFrames.via the plotting module
pandas.plotting.
By default plot() shows a line graph using the row indices a x-axis and the columns as data lines:
airq.Temp.plot()
plt.show()
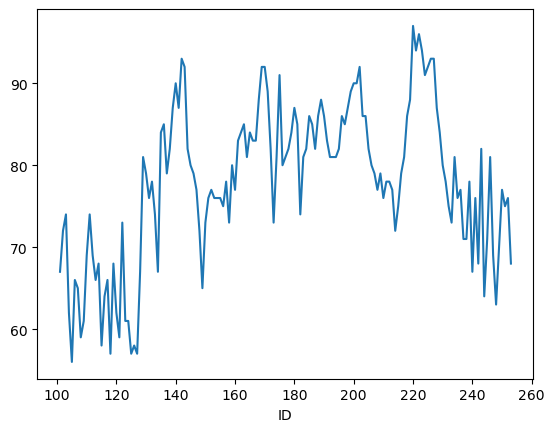
You can create other plots using the methods DataFrame.plot.kind or provide the kind keyword argument to the plot() method:
airq.plot.scatter(x="Ozone", y="Temp", alpha=0.5)
plt.show()

